APTw imaging and segmentation
Project Leaders
Qiuhui Yang
Partner Organisations
National Clinical Research Center for Cancer
Cancer Hospital & Shenzhen Hospital
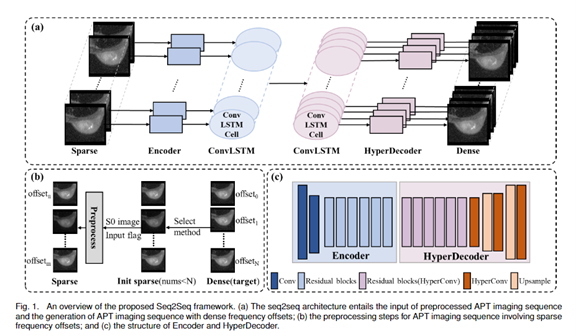
Project Leaders
Qiuhui Yang
Partner Organisations
National Clinical Research Center for Cancer
Cancer Hospital & Shenzhen Hospital

APT fast imaging
Project Leaders
Qiuhui Yang
Partner Organisations
National Clinical Research Center for Cancer
Cancer Hospital & Shenzhen Hospital
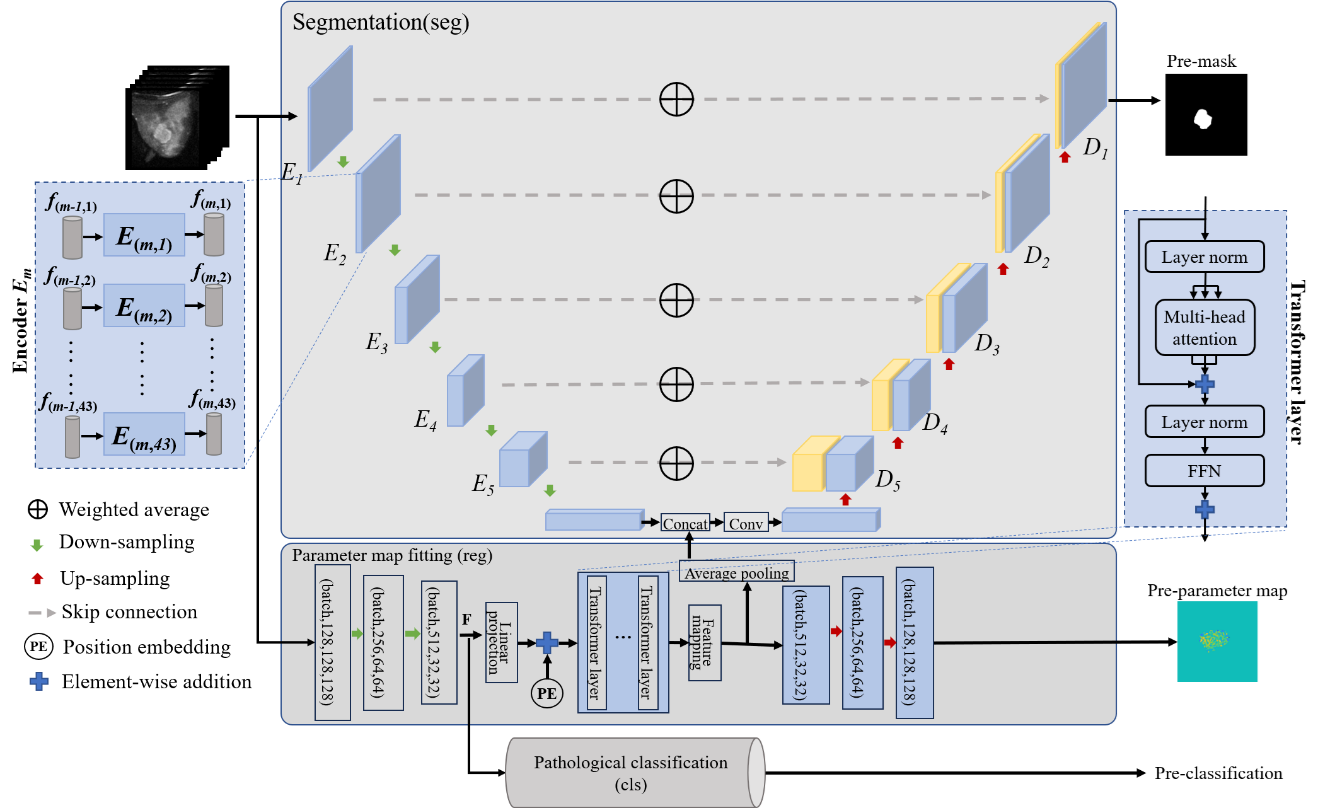
APT Segmentation
Project Leaders
Qiuhui Yang
Partner Organisations
National Clinical Research Center for Cancer
Cancer Hospital & Shenzhen Hospital
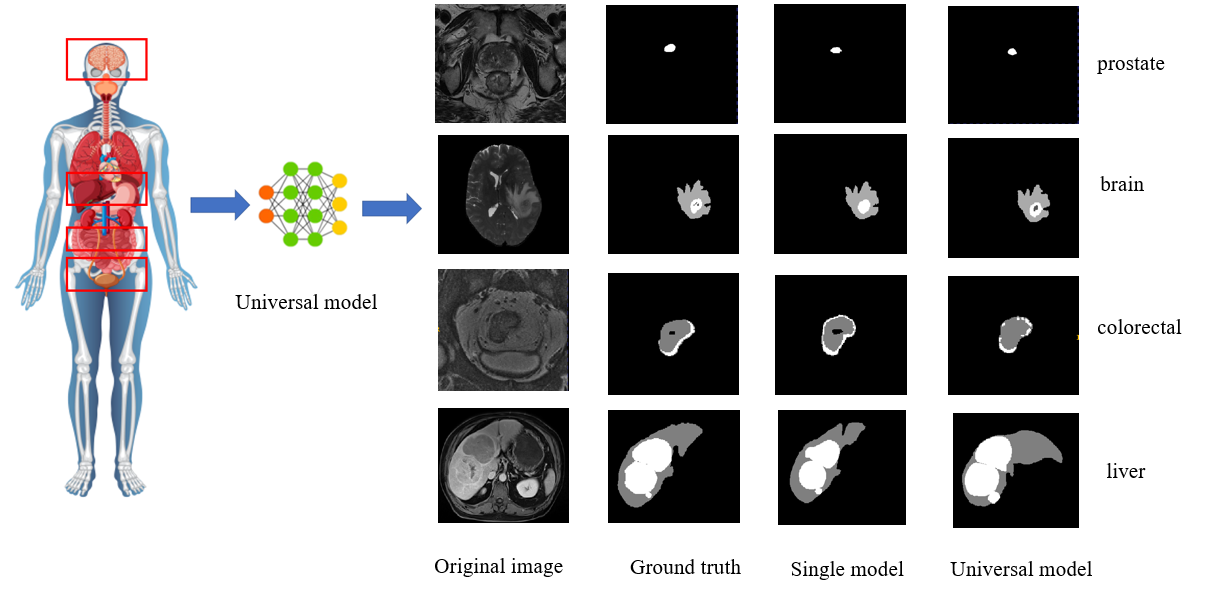
Project Leaders
Mingfu Jiang
Partner Organisations
National Clinical Research Center for Cancer
Cancer Hospital & Shenzhen Hospital
