This research aims to address the problem of information redundancy in multi-sequence magnetic resonance imaging (MRI) to improve the reliability of medical diagnosis. By introducing a sequence-to-sequence generation framework called Seg2Seg, the research objectives include generating 3D/4D sequences arbitrarily, ranking the significance of sequences based on a new metric, and extracting the unique information of each sequence by utilizing the model generation capability.
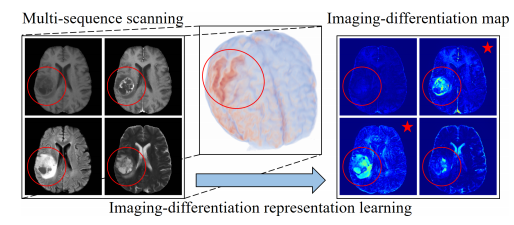
In the field of medical imaging, multi-sequence MRI has been a powerful tool for revealing information within tissues; however, the redundancy of information between sequences has been a challenge that affects the efficient representation of learned models. This challenge poses two major problems: matching of large model training samples is limited, while redundant information between sequences hinders the extraction of diagnostically valuable information. Traditional approaches, such as cropping regions of interest or providing semantic segmentation masks, are employed but still suffer from insufficient global feature association and time-consuming manual labeling. Against this backdrop, researchers Luyi Han, Tao Tan et al, and Ritse Mann (Department of Radiological Sciences, Netherlands Cancer Institute) conducted a study aimed at addressing these challenges by capitalizing on redundancy and imaging disparity information in sequences through an innovative multi-sequence MRI synthesis approach.
The research team introduced the Seg2Seq generator, which enables fast conversion between two arbitrary sequences and successfully extracts the redundant information in each sequence. To cope with the 4D protocol of MRI, the study employed an advanced Long Short-Term Memory (LSTM) mechanism to capture long-term information in 4D MRI. Hyperparameter blocks associated with the target sequence code are used in the network architecture, enabling a single model to generate a variety of sequences. In order to accurately distinguish between imaging differentiation and redundant information, the study generated an imaging differentiation map by comparing the error between the real target sequence and synthetic sequences generated from other sequences.
For Alphabet Recognition, the proposed models Baseline and Baseline+Md were trained, and the results showed an accuracy improvement of 0.262 and 0.369 for X1 and X2, respectively, at a training sample size of 5,000.For MGMT promoter methylation prediction experiments, using the ResNet-18 model, the Comparing the effects of different combinations of input images, the results show that the addition of Md improves the AUC and accuracy, and when all sequences are used, the improvement of AUC reaches 0.023. For breast cancer pCR early prediction experiments, the corresponding Md is used for ResNet-18 training to observe the improvement of the pCR prediction performance, and the results show that in the pre-treatment stage, the model trained with Md in the pCR prediction performance achieved a significant improvement of 0.032 AUC boost in pCR prediction performance.
This study presents an allometric generative model capable of learning imaging difference representations from multi-sequence 3D/4D MRI images to generate missing image sequences. For the first time, the contribution of each sequence in diagnosis is qualitatively revealed, and sequence-specific regions are proposed by imaging difference maps, which can improve the effectiveness of clinical applications such as MGMT promoter methylation status and breast cancer pCR status. The method was demonstrated to be efficient and reliable in the analysis of multi-sequence magnetic resonance images. The researcher, Luyi Han, received a scholarship from the China Scholarship Council (CSC). This research was supported by the Macao Science and Technology Development Fund, and the article was published in "Medical Image Analysis", a top journal of the Chinese Academy of Sciences.
Link to the article:
https://authors.elsevier.com/a/1iBZk4rfPmE0wZ